Research
Research Interests
I am interested in using biophysical tools to understand the protein-protein interactions in the following malaria parasite pathways
Drug-protein interaction of PfGlutathione Reductase
| Taylor Albert | Jan 2023-May 2023 |
| Brielle Skutskie | Jan 2022-May 2022 |
| Alyssa Dam | Jan 2020- May 2020 |
| Anusha Atique | Jan 2019-May 2019 |
| Carolyn Vazquez | August 2017-2018 |
| Robert Mownn | August 2016-2017 |
| Socheata Lim | August 2013-2015 |
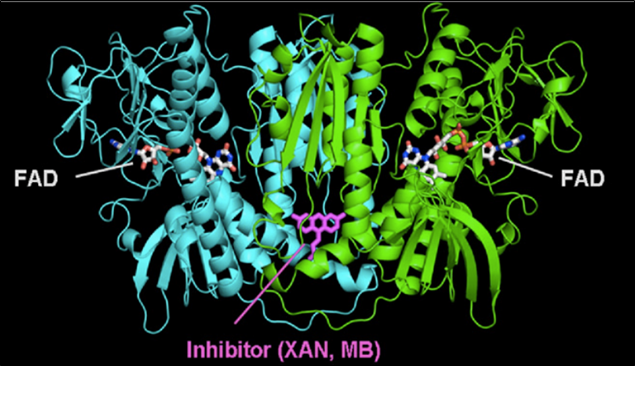 Plasmodium falciparum is the cause of human malaria and is one of two malaria parasites known to have drug resistance. Since there are no preventative vaccinations against malaria, the control of this disease is heavily dependent upon the use of antimalarial drugs. This is especially problematic in underdeveloped countries where the resistance of antimalarial drugs is increasing at a rapid pace. Antimalarial drugs, such as methylene blue, are effective therapies against human malaria. At a specific concentration, methylene blue has been shown to be a selective inhibitor of the parasite’s glutathione reductase (PfGR). Glutathione reductase is an important target when studying malaria drug resistance because it is a flavoenzyme that regenerates glutathione, which is an essential protein for antioxidant defense against cell damage. Methylene blue is also a substrate that is reduced by gluthathione reductase to produce leucoMB. This is then spontaneously oxidized by molecular oxygen to form methylene blue again. During this process, reactive oxygen species, such as hydrogen peroxide and superoxide form. These act as recycling catalysts against infectious organisms. Due to Pf GR’s central position in redox control, it is ranked number one as an antimalarial drug target. Our aim is to characterize the interface between methylene blue and the putative protein target glutathione reductase in order to understand the mechanism of action of the drug. She has expressed and purified Plasmodium falciparum Glutathione Reductase in her previous independent work. Hydrogen-deuterium exchange (HDX) will be the technique used to map the solvent exposed area of the protein and in this way identify the drug-protein interface.
Plasmodium falciparum is the cause of human malaria and is one of two malaria parasites known to have drug resistance. Since there are no preventative vaccinations against malaria, the control of this disease is heavily dependent upon the use of antimalarial drugs. This is especially problematic in underdeveloped countries where the resistance of antimalarial drugs is increasing at a rapid pace. Antimalarial drugs, such as methylene blue, are effective therapies against human malaria. At a specific concentration, methylene blue has been shown to be a selective inhibitor of the parasite’s glutathione reductase (PfGR). Glutathione reductase is an important target when studying malaria drug resistance because it is a flavoenzyme that regenerates glutathione, which is an essential protein for antioxidant defense against cell damage. Methylene blue is also a substrate that is reduced by gluthathione reductase to produce leucoMB. This is then spontaneously oxidized by molecular oxygen to form methylene blue again. During this process, reactive oxygen species, such as hydrogen peroxide and superoxide form. These act as recycling catalysts against infectious organisms. Due to Pf GR’s central position in redox control, it is ranked number one as an antimalarial drug target. Our aim is to characterize the interface between methylene blue and the putative protein target glutathione reductase in order to understand the mechanism of action of the drug. She has expressed and purified Plasmodium falciparum Glutathione Reductase in her previous independent work. Hydrogen-deuterium exchange (HDX) will be the technique used to map the solvent exposed area of the protein and in this way identify the drug-protein interface.
Programmed Cell Death in the malaria parasite
| Kamela Benga | January 2024-May 2024 |
| Xhesika Skoti | September 2023-December 2023 |
| Humayra Meem | January 2023- May 2023 |
| Jack Reardon | September 2021-December 2021 |
| John Carroll | January 2020-May 2020 |
| Max Simpson | January 2018-May 2019 |
| Zena Wright | August 2017-May 2017 |
| Carlos Cardenas | August 2017-Dec 2017 |
| Donesiha Coleman | August 2016-May 2017 |
| M. Hazal Yilmaz | August 2016-2017 |
| Erick Orozco- Morato | August 2014-Current |
| Maha Ali | Summer 2014 |
| Amrita Bains | August 2014-May 2015 |
| Nicholas Darinzo | August 2013-May 2014 |
| Patrick Finneran | January 2013-May 2014 |
| Thaiani Paca | January 2013-May 2013 |
 Project Programmed Cell Death is one of the control mechanisms, used by higher multicellular animals that are crucial for the continuing development of the surviving cells. Recently more reports have identified a similar mechanism in unicellular organisms and the pathways are starting to be identified. It was not expected that a protozoan parasite such as the causing agent of malaria would show signs of this programmed “suicide”. Our research interest lies in establishing the existence of this pathway in the erythrocytic stages of the parasite. Apoptosis is regulated by cytochrome c and a downstream protease in all mechanisms known to date. Previous students have worked to develop an assay to test the role of suspected proteases in the activation of an apoptosis pathway using yeast as a model organism. Understanding of this pathway in yeast leads to test the hypothesis of a similar protease involved in P. falciparum. Our aim is to establish the assay first in yeast and subsequently in Plasmodium to test cytochrome c’s activation of a peptidase by using fluorescence spectroscopy and a peptide substrate.
Project Programmed Cell Death is one of the control mechanisms, used by higher multicellular animals that are crucial for the continuing development of the surviving cells. Recently more reports have identified a similar mechanism in unicellular organisms and the pathways are starting to be identified. It was not expected that a protozoan parasite such as the causing agent of malaria would show signs of this programmed “suicide”. Our research interest lies in establishing the existence of this pathway in the erythrocytic stages of the parasite. Apoptosis is regulated by cytochrome c and a downstream protease in all mechanisms known to date. Previous students have worked to develop an assay to test the role of suspected proteases in the activation of an apoptosis pathway using yeast as a model organism. Understanding of this pathway in yeast leads to test the hypothesis of a similar protease involved in P. falciparum. Our aim is to establish the assay first in yeast and subsequently in Plasmodium to test cytochrome c’s activation of a peptidase by using fluorescence spectroscopy and a peptide substrate.
The parasite lacks the usual protease expected in metazoan, a caspase. A large upregulation of the dipeptidyl aminopeptidase-1, DPAP-1, was detected in a quantitative proteomic study where the parasite was subjected to treatment with antimalarial drugs. DPAPs are proteases recognized to have a function in hemoglobin degradation in the erythrocytic stages of the malaria parasite, Plasmodium falciparum. DPAP-1 has been compared to the human ortholog cathepsin C in previous studies, concluding that both proteases belong to the same family of C-1 endo- and exopeptidases. We are interested in finding if DPAP-1 is the protease downstream of cytochrome c.
Characterization of the Folate Pathway in Malaria
| Samantha Burns | January 2022-May 2022 |
| Francis Astudillo | January 2021-May 2021 |
| Mariah Nieves | September 2020-December 2020 |
| Alyssa Dam | Fall 2018-May 2019 |
| Layra Valdes | Fall 2017-May 2018 |
| Emily Federowicz | Summer 2017 |
| Sumra Akhlaq | August 2016-May 2017 |
| Karen Anderson | August 2014-June 2015 |
| Carlos Menjivar | August 2013-May 2014 |
| Sarah Castro | August 2014-Current |
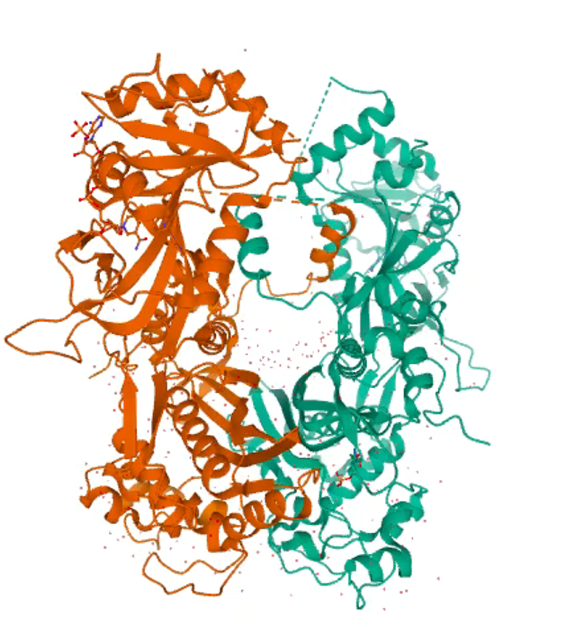
Dihydrofolate reductase (DHFR) is an integral enzyme to many biological systems, catalyzing the conversion of dihydrofolate to tetrahydrofolate (THF), which in turn is required for the synthesis of nucleic acids. DHFR is well known target for cancer and malaria as a lack of nucleic acids synthesis interferes with cell growth and proliferation. Sulfadoxine (SP) and pyrimethamine (PYR) were both drugs that targeted the folate pathway in malaria efficiently until resistance spread throughout the world. Recent research describes the way mutations were able to change the conformation of the enzyme to decrease its binding affinity for the inhibitors, resulting in the resistance of the enzyme to structural analogs such as SP or PYR. Notably once the drug pressure is taken off the field the parasite favors the expression only of the wild type protein. We aim to understand with this project how the conformation of the mutants affects the structure as well as the function of the protein.